Examples
- Introduction
- Designing padlock probes
- Designing minisequencing primers
- Calculating Tm for a set of sequences
Introduction
This file will provide a number of examples on how to use ProbeMaker for different oligonucleotide design and analysis tasks. Hopefully, the number of examples provided will increase as new uses of the software are reported.
If you have questions about the use of ProbeMaker, or need an addition to the software framework, visit the sourceForge project site and submit a support request/bug report/request for enhancement, or send an email to the probemaker-users mailing list.
If you haven't already, it would be a good idea to go through the ProbeMaker tutorial.
Designing padlock probes
An example of how to perform padlock probe design can be found in the tutorial.
Designing minisequencing primers
To design a set of minisequencing primers, perform the sequence input as in the previous example, but using the 'minisequencing target' target type rather than padlock probe targets.
[Unfortunately, the input modifier that is supposed to select the target SNP does not handle minisequencing targets correctly (working on a fix!), so the targets that contain multiple SNPs will have their first SNP selected rather than the one intended. To fix this, manually set the variant to SNP 2+ for the targets rs1288699, rs4952450, rs4983727, and rs6047983.]
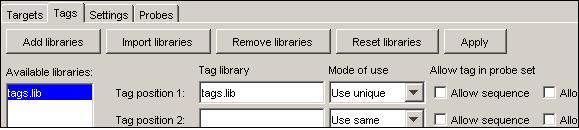
Use the 'Expand targets' button to create a target for each strand if desired. The minisequencing targets will generate a single (3') target-specific sequence, ending adjacent to the SNP site on the target strand. To generate a tagged minisequencing primer for tag array readout experiments, a unique tag sequence needs to be added to the 5' end of each primer. To setup ProbeMaker for this, load the tag library ('tags.lib') and add it to the first tag position. Set the mode to 'Use unique' in order to use add a unique tag to each probe (see figure).
Click the 'Apply' button and the design job is ready to be run. This will create a set of tagged primers. However, the selection mechanism has not been used (nor was it used in the previous example). To try this out, enter the 'Settings' tab and drag and drop analysis modules from the list of available modules to the different evaluation stages. In this way, allocate the 'TSS length and temperature' module to the TSS evaluation stage. This will display the 4 tests this module provides. Un-check the two boxes that regard tests of the 5' TSS, as the probes we are making don't have 5' TSSs. Also, check out what parameters this module has by double-clicking its name. This module allow the setting of a cut-off mode. Using the cutoff mode means that TSSs with a Tm above the preferred one will be allowed (click the module name in the left-hand list for a detailed description). Allocate the 'False priming' module to the TSS stage as well. This will chack all primer for the risk of false priming on other primers or targets according to the algorithm by Kaderali et al., as summarized in the module description. Now run this project, using the 'Accept good candidates, fair ones for selection' acceptor, and the 'select probe with best quality' selector. This should result in that a number of probes fail, mostly because they risk being primed by other targets in the target set. The tags are still selected in order from the provided list, since the choice of tag does not help this problem. Add the 'Tm-based probe dimer formation module' to the stage 1 evaluation box, and run a new design (don't forget to click 'Apply'). This will take a few minutes to complete (although you can abort the task and view partial results), as each candidate probe is compared to all other probes to estimate the risk for probe-probe dimer formation (according to the Tm algorithm by Leber et al.). When it is finished, bring up the tag allocation table ('view tags by probe'). Now you will see that some tags have not been used, or have been used in a different order. This is because some primer-tag combinations increase the risk of probe homo- or hetero-dimer formation.
Calulating Tm for a set of sequences
The ProbeMaker framework can be used for other tasks than probe-design as well. An example is to calculate the melting temperature of each of a set of sequences. This can be done by utilizing the TSS design function that computes the Tm. The Tm can be properly calculated only for sequences without polymorphisms. So, let's load all the tag sequences and calculate their Tm. Import the file 'tags.lib' as 'Whole sequence as template' type targets, in Fasta format and press cancel when prompted to select converter and modifier, as this is not needed. On the 'tags' tab, remove any allocated tag libraries by clicking in the tag position text field. Click the apply button before running. To view the Tm results, click the 'Probe summary' button on the 'Probes' tab, or use the corresponding menu option.

