Tutorial
- Introduction
- Setup
- Target input
- Setting design parameters
- Running a design job
- Looking at the results
- Creating a project template
- Troubleshooting
Introduction
This tutorial will guide you through setting up the ProbeMaker software, creating a project template and running a design job. It will also provide some basic knowledge of how ProbeMaker works and the functionality available.
The ProbeMaker tutorial makes the following assumptions:
- Your computer has a Java Runtime Environment of at least version 1.4 installed (If not, visit http://java.com)
- You have a ‘probemakerX.Xzip’ or ‘apptools-X.Xzip’ (or similar) file downloaded from http://sourceforge.net/projects/probemaker
- You are running Microsoft Windows (The tutorial is valid even if not, but some details may differ)
If you encounter any problems, please see the troubleshooting guide before submitting a support request through the sourceforge.net project site.
Setup
Step 1, Unpack:
Start by unpacking the downloaded file in a convenient location on your system. From version 1.2 on, ProbeMaker runs through the so-called AppTools workbench. The zip-file distribution of ProbeMaker contains this workbench and other required components. Unpacking the zip file will create a directory named apptools with a number of subdirectories on your file system.
Note that if the Windows compressed files utility is being used, you can view these files without having extracted them from the archive. You need to extract them for ProbeMaker to function properly. Use the ‘Extract all files’ option.
Step 2, Launch the AppTools workbench:
Double-click the ‘start.bat’ file to start the AppTools workbench (if this does not work, Java has not been properly set up, see the troubleshooting guide for help).
The autoact.txt file in the plugins directory contains a list of all the extensions that should be activated at start-up. If this file is empty, plug-ins may be loaded via the Options->Activate plugins dialog.
Step 3, Activate the ProbeMaker plug-ins:
Open the 'activate plugins' dialog using the Options->Activate Plug-ins menu option. Choose to load the ProbeMaker GUI plugin.
Target input
Step 4, Creating a new project:
Create a new project by clicking the ‘New project’ icon or the File->New project menu option. Enter a project name and press ‘OK’.
This will create your new project. You will learn about project templates later. You will now import a target sequence file to design a set of padlock probes. The data in the file was once downloaded with SNPper and is in text table format, with target SNPs denoted by [X], where X is the appropriate degeneracy code. There are also other variations in some of the sequences, denoted by ‘N’.
Step 5, Importing target sequences:
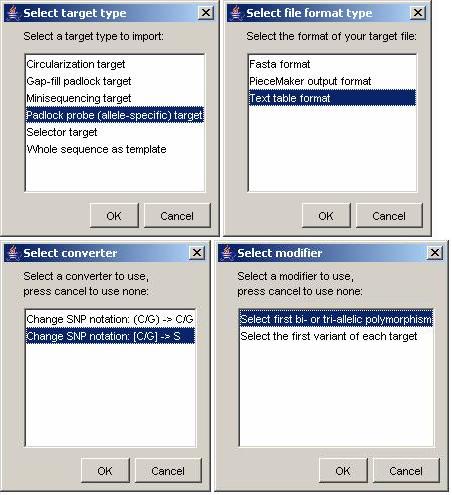
Press the ‘Import targets’ button to read target sequences from file. You will be prompted for four selections and then to select a file.
- First select padlock probe targets, as that’s what we are making at the moment.
- Then select the text table format.
- Since the SNPs are enclosed by brackets, which are not supported by ProbeMaker, you need to use a sequence converter. Select the [C/G] -> S converter (this will remove the brackets around SNPs for both the C/G and S-style notations).
- Since some of the sequences have multiple variations, and you don’t want to select the target SNP for every polymorphism manually, use the ‘select first bi- or tri-allelic polymorphism’ modifier. This will do the trick for you.
- After you’ve done this, you’ll be asked to select the file to load. Select the ‘SNP targets.txt’ file.
This should load 50 SNP-containing padlock probe targets that will be shown on the target tab. You will notice that some have SNP 1 selected, while some have SNP 2. This is because the modifier you used will select the first SNP with two or three variants, skipping those with four (the N’s). This procedure is a bit lengthy, and can be simplified by creating a project template. You will learn about that later on.
Step 6, Expanding targets:
Now, there is one sequence per target, but we actually want to make two probes for each target, so press the ‘Expand’ button to create two targets for each sequence. These will differ only in which SNP allele is selected, and will be grouped together to allow locus-specific tags to be added.
Hint: You can access more information about a target sequence by right-clicking an entry in the target table.
Setting tag libraries
We are making padlock probes, so we will add two primer sequences and an address tag (for microarray hybridization) to each probe. One primer should be common to all probes, while the other primer is either of two allele-specific sequences. The address tag should be same for each pair of probe, but different for different pairs. These settings are made on the ‘Tags’ panel by importing and allocating tag libraries to different positions and setting their selection mode.
Step 7, Importing tags:
For this tutorial, we provide three files with tag sequences:
- common_primer.lib
- allele_primers.lib
- tags.lib
Press the ‘Import libraries’ button and select these three files. Then select the Fasta format from the format dialog. The three libraries will now be displayed in the libraries list.
Hint: You can get more information by moving the mouse pointer over the library names and by clicking them.
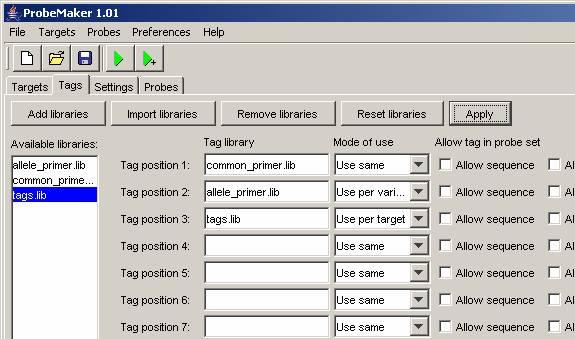
Step 8, Setting tag usage:
Drag and drop the tag library names to the desired positions and set the ‘Mode of use’ as shown in the figure below. Then press the ‘Apply’ button to apply the changes to the current project.
Setting design parameters
On the Settings tab a number of parameters affecting the design job can be set. Also, this is where analysis modules are allocated to perform tests on the generated probe candidate sequences.
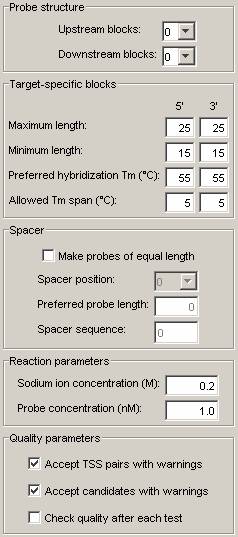
Step 8, Setting design parameters:
Go to the Settings tab and check that the settings are as shown in the figure below. If they are not, the default settings file has probably not been correctly set in step 2. In such case, fix the parameters and go on. You can go back and set the parameters file later.
Analysis modules are what provide the ProbeMaker framework with the ability to analyze probe and target sequences. Each analysis module provides one or more tests, and may provide a set of parameters to affect how these tasks are carried out. Analysis modules may be allocated to three stages; TSS testing, candidate evaluation stage 1, and candidate evaluation stage 2. The first stage is performed after all target-specific sequences have been designed, and is suitable for tests that need to be carried out only once per probe. The other two stages are carried out for each candidate of a probe (each valid combination of tag sequences), and the results of stage 1 evaluated before stage 2 is carried out (so time-consuming tests should be put in stage 2).
Analysis modules are plug-in extensions, so new modules providing new tests can be constructed and easily incorporated into the ProbeMaker framework.
Step 9, Allocating analysis modules
Drag and drop the module named ‘TSS length and temperature module’ to the TSS testing box (the left box of the top right pair of boxes). If your screen resolution is small, you might need to use the scrollbar at the bottom of the ProbeMaker window to see this box completely. Four tests will now be displayed in the corresponding test box on the far right. The checkboxes are used to set which tests to carry out. By double-clicking the module name in the testing box, the parameters for this module will appear and may be set. Leave the settings unchanged. Now press the ‘Apply’ button to apply these changes to the current project.
Hint: You can get a description of what each module does by double-clicking its name in the list of available modules.
Running a design job
Everything is now set to perform your first design job. Before this, it might be a good idea to save your project. Press the save icon or select the File->Save project menu option to bring up a save dialog.
Step 10, Running a design job
Press the ‘Design probes to replace’ icon in the tool bar (the green triangle without the plus sign). You will now be asked to select an acceptor and a selector, and if you wish to reset the tag libraries. Select the ‘Accept good candidates, Fair ones for selection’ acceptor, and the ‘Select probe with best quality’ selector. [Version 1.2 also allows you to choose any of a number of 'TSS constructors' that determine how the target-specific sequences of the probes are constructed. For now, choose the one named 'Target-complementary sequences'] Press ‘OK’ and the design job will start.
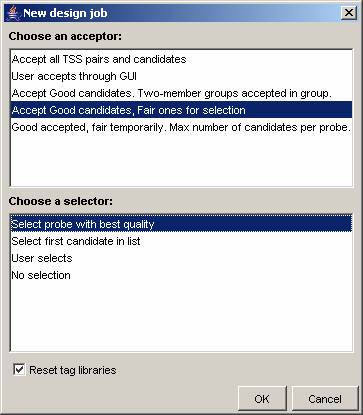
The design job should be completed in about a second or two, and the view will change to display the newly designed probe sequences. Most will be displayed in green, meaning that they are of the highest quality (no warnings or errors attached to these probes), while some will be displayed in yellow, meaning that they are of the intermediate quality level (one or more warnings attached). You can bring up detailed probe information by right-clicking each probe. As you will see, the warnings are all about probe melting temperature. The maximum length of 25 bases was not enough to reach the temperature range of 50 – 60 °C for some very AT-rich target sequences.
Looking at the results
There are a few different ways to display results of the design. Try some or all of the actions in the list below to explore this:
- Probe summary: press the ‘Probe summary’ button to view a list of all probes, their 5’ and 3’ Tm values, and their quality value.
- Message summary: press the ‘Message summary’ button to view a list of the types of problems that have occurred, and their frequency.
- Tag listing: press the ‘View tags by probe’ button to display a table of which tag has been allocated to which probe. (Since the tests used in this design job do not affect the selection of tags, all tags will be in the order they were in the library file)
- Probe export: The ‘Export probes’ button allows the probes to be exported to file in a variety of formats, and new such formats can be created and plugged-in.
Hint: If your screen resolution is low, or your ProbeMaker window is small, some of these buttons may not be visible. The same options are available from the 'Probes' menu.
You how now completed a project. Try out some of the other analysis modules to see how they affect the design (e.g. Padlock TSS ligation, which should also go in the TSS testing stage), or go on to learn about project templates.
Creating a project template
The target input system allows plug-ins to be created for most conceivable sequence formats and target types. However, the procedure of selecting four plug-ins each time may be a bit cumbersome. The same is true for selecting design parameters and analysis modules. The project template system allows the creation of new projects based on a template that defines the input method used and sets parameters and analysis modules to a default setting for a particular type of design. You will now learn how to create a project template for the padlock design you have carried out.
Step 11, Creating a project template:
First, go to the settings panel and press the ‘Save parameters’ button to save your current settings to a file named ‘my settings.par’.
Then, select the Preferences->Project templates menu option to bring up the templates dialog. Create a new template with the ‘New template’ button and select the template components as you did for the target input in step 5. Select your newly created ‘my settings.par’ as settings file.
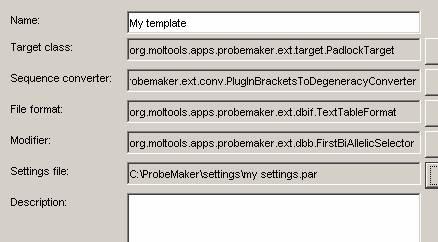
Hint: Fill out the description field if you want to. If the description is long, consider formatting it in html by enclosing it with html tags. (e.g. ‘<html>Template for <b>Padlock probe</b> design using <i>text table</i> target format</html>’.
Press the ‘OK’ button when you are done. To test your template, create a new project (you will be asked whether you want to save your current project). In the new project dialog, make sure to check the ‘Use template’ box and to select your new template before pressing OK.
When you import targets with your new project, you will not have to specify import plug-ins. Also, the settings file is loaded to provide the same parameters and analysis module settings for each project created with the same template. Thus, ProbeMaker can be tailored for your particular requirements.
Troubleshooting
ProbeMaker does not start when the icon/file is clicked.
This is probably due to Java not being installed properly, not at all, or that the Java version is too old.
First, check that your operating system knows where your java interpreter (java.exe) is. This can be checked quickly as follows;
- Selecting the ‘Run…’ item on the Windows start menu.
- Type ‘cmd’ in the field and press OK. This starts the windows command interpreter.
- Type ‘java -version’ at the command prompt. If a version
message is displayed, you’re OK. If an error message is
displayed instead, the ‘PATH’ environment variable may be
set to include the path of the java interpreter, as follows (Windows
XP):
- On the start menu, select Settings > Control Panel.
- Locate and open the ‘System’ settings. In ‘Category View’ this is found under ‘Performance and Maintenance’.
- Select the ‘Advanced’ tab, and click the ‘Environment Variables’ button.
- Locate the System variable ‘Path’ and edit it.
- Add the path of your java interpreter (e.g. ‘c:\jre1.5.0_02\bin’) to the value. Use a semicolon to separate path entries.
- Repeat the test procedure above to see if it worked.
The above procedure also gives you the version of Java installed. Version 1.4.2_07 or later is recommended, as this was used when developing ProbeMaker.
Now, still in the command window, move to the directory where you installed ProbeMaker by typing: ‘cd \ProbeMaker’. Then type ‘java –jar ProbeMaker.jar. This should now start the ProbeMaker program.
I can’t find the ProbeMaker directory and files when I’m setting the file preferences.
Make sure you have actually unpacked the ProbeMaker.zip file and are not running the program from within the archive.
I had some trouble the first time I started ProbeMaker and now I am not asked to set the file preferences when I start the program, I just get some error messages instead.
The file settings are saved to the system when you set them, so they will stay even if you remove the program. Click past the error messages and bring up the User preferences dialog via the Preferences->User preferences menu option.

